Plotting occurrence data from the query "Download record counts of Peron's tree frog since 2018 in New South Wales by FrogID"
Overview
Teaching: 10 min
Exercises: 10 minQuestions
How can I make a map using occurrence records from the ALA?
Objectives
Be able to download occurrence records, including select columns of data
Plot a shape file using geopandas
Plot occurrence records from the ALA in context with a shape file
Download occurrence records
Remember the query we built from Episode 2:
Download record counts of Peron's tree frog since 2018 in New South Wales by FrogID
galah.atlas_counts(
taxa="litoria peronii",
filters=["year>=2018",
"cl22=New South Wales",
"dataResourceName=FrogID"]
)
totalRecords
0 39840
All we have to do to download occurrences is to change the function name atlas_counts to atlas_occurrences, but first, we need to provide an email registered with the ALA to galah-python:
galah.galah_config(email="amanda.buyan@csiro.au")
galah.atlas_occurrences(
taxa="litoria peronii",
filters=["year>=2018",
"cl22=New South Wales",
"dataResourceName=FrogID"]
)
decimalLatitude decimalLongitude eventDate ... recordID dataResourceName occurrenceStatus
0 -37.246800 149.375000 2020-12-27T00:00:00Z ... 56de6c10-b14d-4eeb-86ee-a50e406678d3 FrogID PRESENT
1 -37.245410 149.957044 2022-10-26T00:00:00Z ... 685271bf-5bbf-4174-b8e2-c92c4b5ee586 FrogID PRESENT
2 -37.245385 149.957663 2021-12-11T00:00:00Z ... 4155af9f-38bf-4093-976f-65d17029d1c4 FrogID PRESENT
3 -37.245385 149.957663 2021-12-11T00:00:00Z ... aee93398-6e27-4134-a1c3-9a4c794d1ce6 FrogID PRESENT
4 -37.245385 149.957663 2021-12-11T00:00:00Z ... 360887c3-4af9-4430-801e-8b7db47a2389 FrogID PRESENT
... ... ... ... ... ... ... ...
39835 -28.207514 153.442592 2018-11-15T00:00:00Z ... 10ec5c96-fb1e-4545-9e85-dc9073e3a977 FrogID PRESENT
39836 -28.207494 153.442526 2021-11-17T00:00:00Z ... 302250c5-6b7b-4d5d-b51b-12fa305ae8c9 FrogID PRESENT
39837 -28.207472 153.442497 2018-11-15T00:00:00Z ... 101f5f04-b0e9-45b4-a9c0-3e50d97f1dfe FrogID PRESENT
39838 -28.207442 153.442328 2020-02-07T00:00:00Z ... f827c2ef-fcf4-40cc-ab3d-fc1f4ec8c61b FrogID PRESENT
39839 -28.207108 153.443021 2021-02-19T00:00:00Z ... f02823d8-f53a-4cde-b546-d3bd7ff7b075 FrogID PRESENT
All of this data for each occurrence record is great! However, say you want to only get specific columns of the table, like decimalLatitude,decimalLongitude and scientificName. You can specify column names in the fields argument of atlas_occurrences:
galah.galah_config(email="amanda.buyan@csiro.au")
galah.atlas_occurrences(
taxa="litoria peronii",
filters=["year>=2018",
"cl22=New South Wales",
"dataResourceName=FrogID"],
fields=["scientificName","decimalLatitude","decimalLongitude"]
)
scientificName decimalLatitude decimalLongitude
0 Litoria peronii -33.624100 151.323000
1 Litoria peronii -33.718800 151.003000
2 Litoria peronii -33.324700 151.365000
3 Litoria peronii -33.572700 148.436000
4 Litoria peronii -35.115900 147.981000
... ... ... ...
39835 Litoria peronii -33.817474 151.177367
39836 Litoria peronii -33.948932 151.251668
39837 Litoria peronii -33.930552 151.237679
39838 Litoria peronii -33.686587 151.096895
39839 Litoria peronii -33.448529 151.375129
Make a map of Litoria peronii occurrence records since 2018 in New South Wales
Now, we will make a map showing where all the occurrence records we downloaded are in New South Wales. This is where the packages matplotlib and geopandas you installed at the beginning of the lesson will be used. The shape file you will need for this exercise can be found here.
import galah
from matplotlib import pyplot as plt
import geopandas as gpd
# Get Peron's tree frog occurrences
frogs = galah.atlas_occurrences(
taxa="litoria peronii",
filters=["year>=2018",
"cl22=New South Wales",
"dataResourceName=FrogID"],
fields=["scientificName","decimalLongitude","decimalLatitude"]
)
# Get Australian state and territory boundaries
states = gpd.read_file("STE_2021_AUST_GDA94.shp")
# Change Coordinate Reference System (CRS) of the shape file and plot New South Wales
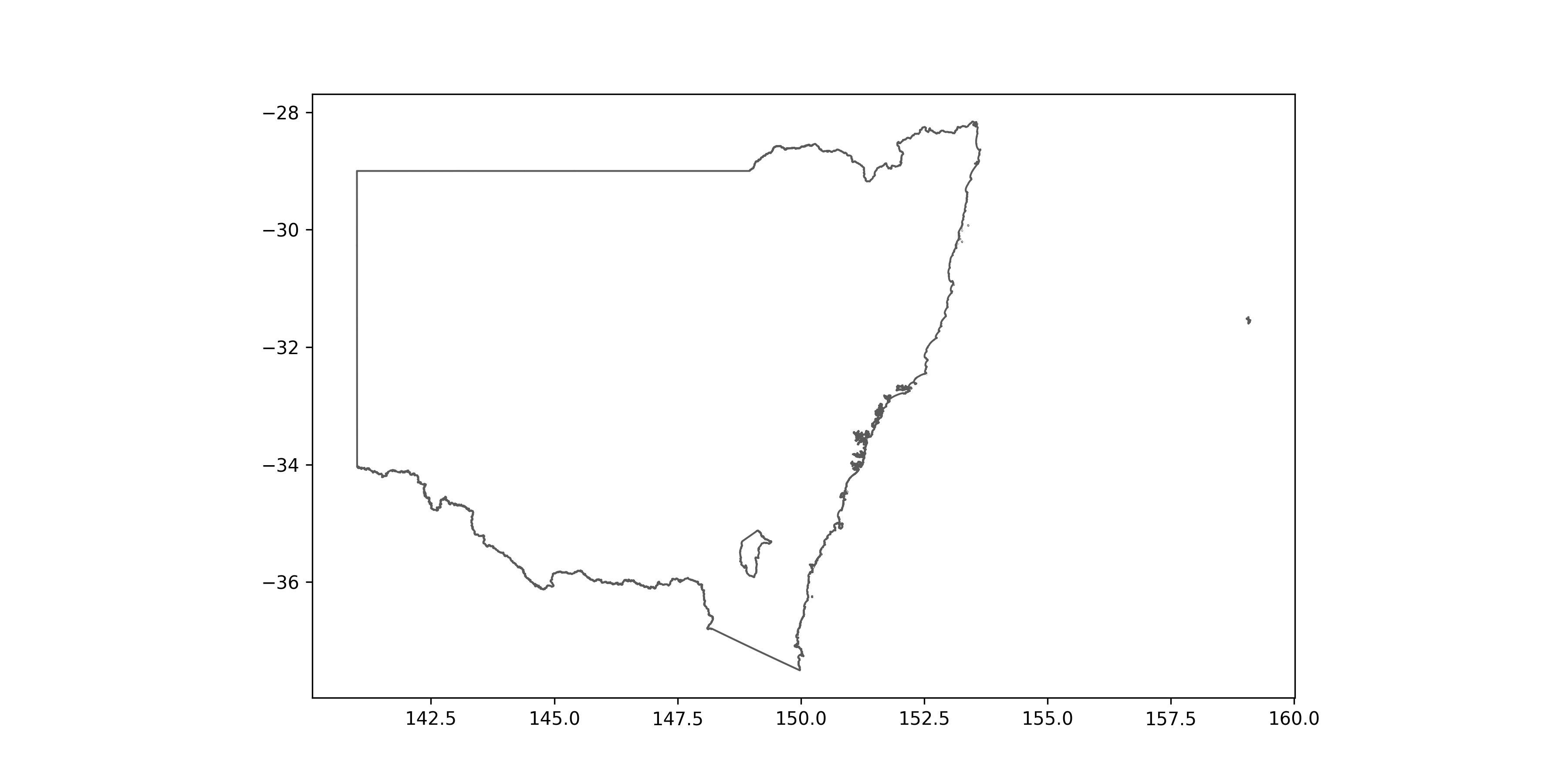
states = states.to_crs(4326)
states[states["STE_NAME21"] == "New South Wales"].plot(edgecolor = "#5A5A5A", linewidth = 1.0, facecolor = "white", figsize = (24,10))
# Add occurrence records to the map
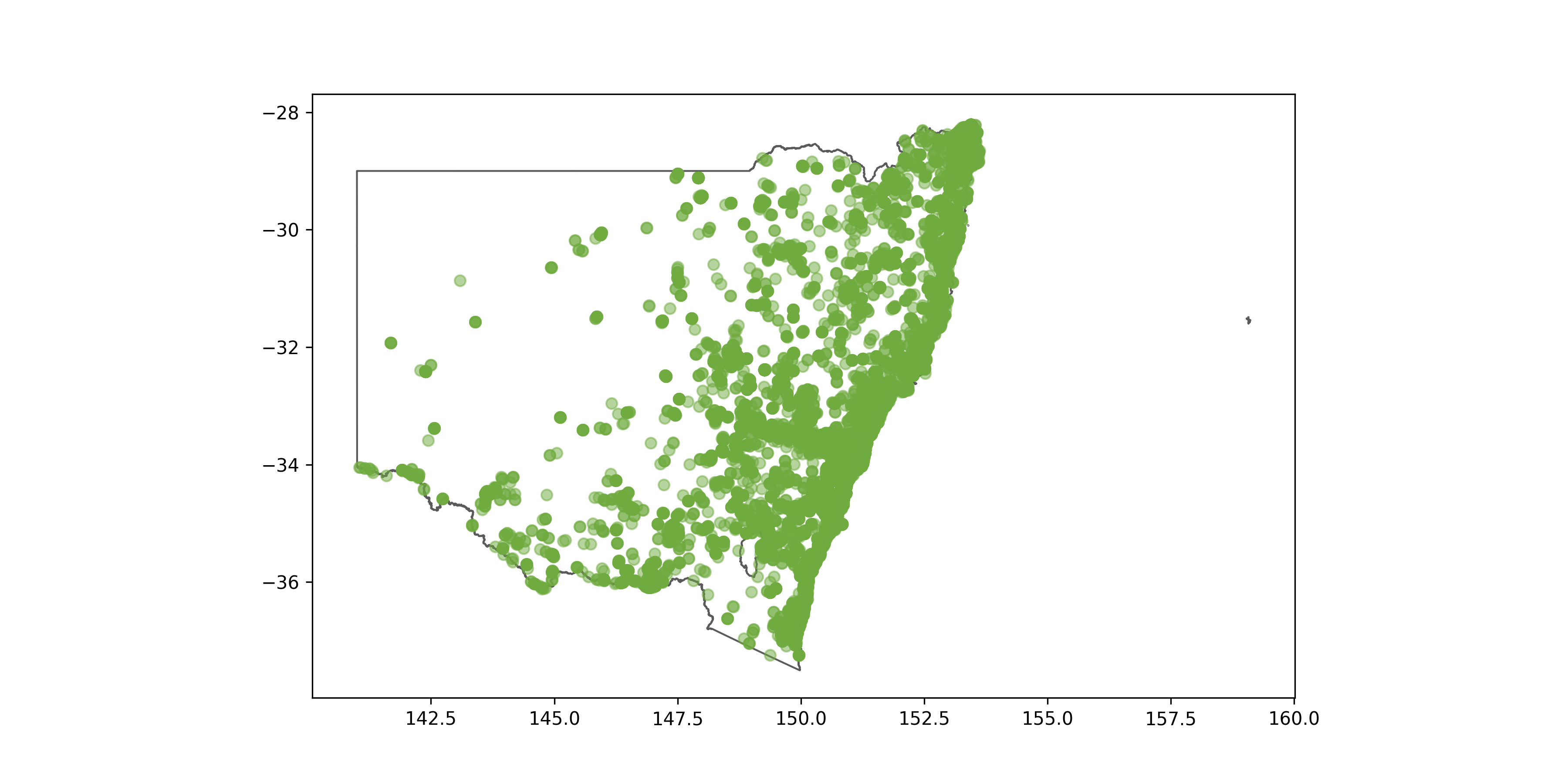
ax = states[states["STE_NAME21"] == "New South Wales"].plot(edgecolor = "#5A5A5A", linewidth = 1.0, facecolor = "white", figsize = (24,10))
plt.scatter(frogs['decimalLongitude'],frogs['decimalLatitude'], c = "#6fab3f", alpha = 0.5)
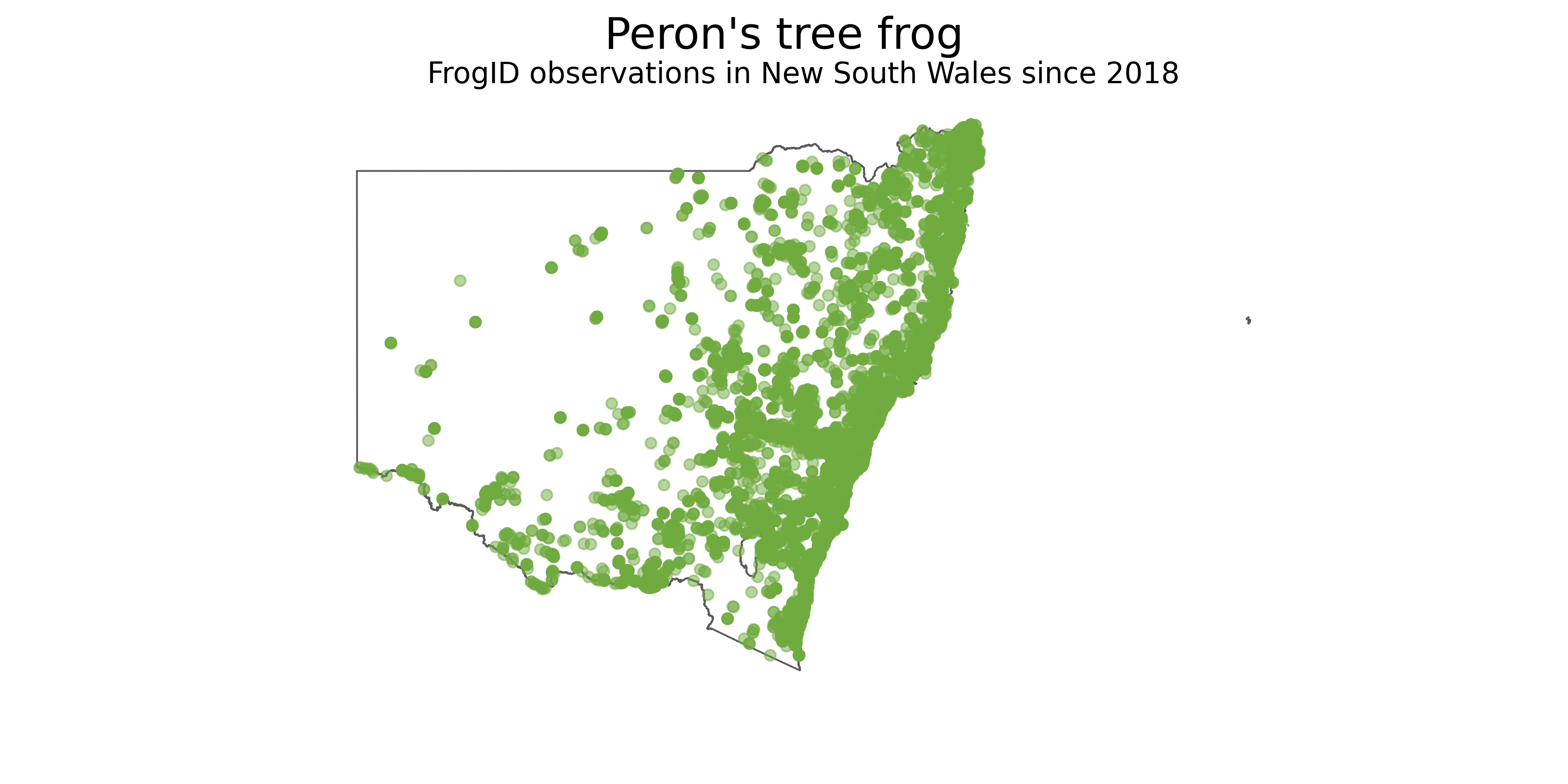
# Add final touches to figure
plt.suptitle("Peron's tree frog",fontsize=24)
plt.title("FrogID observations in New South Wales since 2018",fontsize=16)
plt.axis('off')
Key Points
Occurrence records can be downloaded by using queries from
atlas_counts(), but using the functionatlas_occurrencesinsteadYou can specify what columns of data you want from
atlas_occurrences()by using thefieldsoptionPlotting the occurrence data on a map can be done with
galah-python,matplotlibandgeopandas